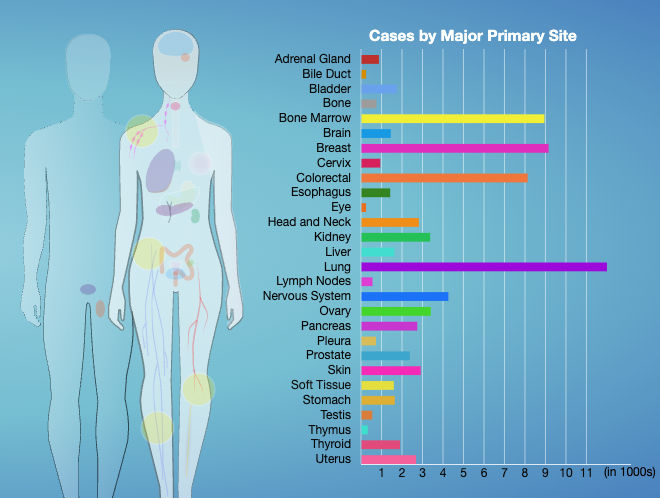
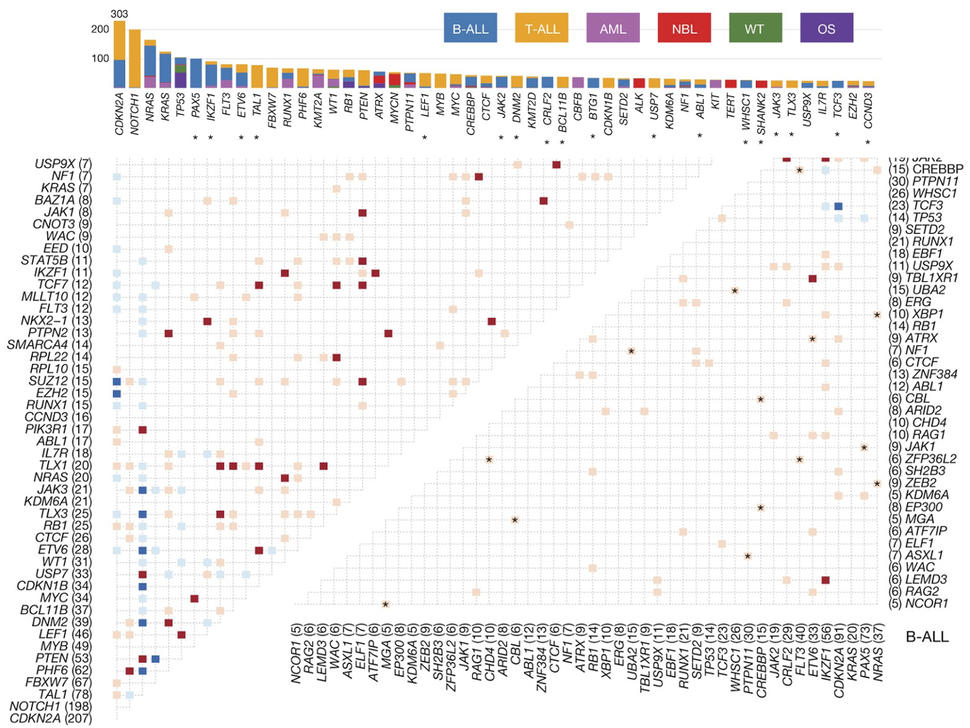
Therapeutically Applicable Research to Generate Effective Treatments (TARGET) researchers used various sequencing and array-based methods to examine the genomes, transcriptomes, and, for some diseases, epigenomes of select childhood cancers. This “multi-omic” approach generated comprehensive profiles of molecular alterations for each cancer type.
Robust clinical data were also obtained for each case studied in TARGET. All tissues used meet strict scientific, technical, and ethical requirements. Selection of patient cohorts were based upon characteristics specific to each disease or cancer subtype.
All data collected and processed by the TARGET program is available at the Genomic Data Commons (GDC):
Access Primary Data at the GDC Data Portal |
Find Analysis Files from TARGET Publications in the “Supplemental Data” Section of GDC Publication Pages |
Data for each project was generated in two phases:
- Discovery: Characterization of a “discovery” cohort of patient cases is used to identify molecular alterations of the transcriptome, genome, and epigenome. For each case, tumor tissue taken at the time of diagnosis, case-matched “normal” tissue, and whenever available, case-matched tissues from relapsed or treatment-resistant tumors were characterized.
- Validation: An independent cohort of patients were analyzed via a separate sequencing method to validate at the gene level most candidate mutations found in the discovery phase. Validation confirmed the population prevalence of mutations. Within each disease, characterizing a separate cohort more broadly representative of the disease population further allowed the project teams to estimate the frequency of somatic variants in a given cancer subtype.
Learn more about how the data was generated and other details important for using TARGET data and other related resources in your research: