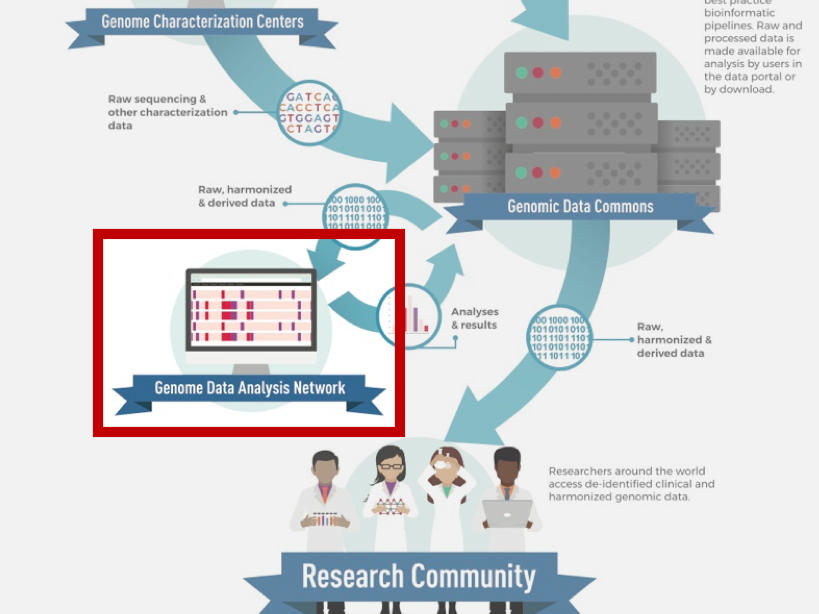
About the Genomic Data Analysis Network
While large genomic datasets are invaluable to the cancer research community, translating genomic data into biological insights into the development and treatment of cancer is not a straightforward task. Over a decade of experience from The Cancer Genome Atlas (TCGA) program demonstrated the power and necessity of “team science”—that successful analyses of large-scale genomic datasets require the coordination of a large body of researchers with a wide range of expertise in computational genomics, tumor biology, and clinical oncology.
OCG’s Genomic Data Analysis Network (GDAN) was formed from the need to harness TCGA data and a growing need at large for computational genomics. For TCGA, the network created standardized data formats and processing protocols, generated bioinformatics tools for the community, and performed a range of analyses on the data, notably generating clinically meaningful molecular subgroups of cancer and producing the PanCancer Atlas.
In the post-TCGA era, the GDAN continues to conduct key large-scale studies and generate genomic resources to support the genomic research community. The GDAN’s overall goal is to help the cancer research community leverage the genomic data and resources produced by OCG and other NCI programs for the benefit of cancer patients, largely by:
- developing and implementing new bioinformatic and computational tools to capture key biological insights about cancer (e.g., pathway analysis, data integration with visualization, and integrated cancer biology);
- developing data processing and quality control methods for working with large-scale genomic characterization data;
- processing and integrating a variety of analytical data types to generate disease-level findings and perform cross-disease analyses.
The GDAN is comprised of individual Genome Data Analysis Centers (GDACs), each specializing in a unique set of computational analyses, molecular platforms, data integration, or visualization techniques. The GDACs are tasked to cooperatively perform molecular analyses on new and existing data from OCG programs and work with the other components of OCG’s Genome Characterization Pipeline. Areas of expertise and examples of their utility include:
- DNA Mutations – Identifying mutations in coding and non-coding regions of the genome, classifying mutations as driver or passenger mutations, identifying chromosomal rearrangement events leading to fusion proteins, and determining potential enhancer or suppressor functionality of mutations.
- Gene Expression – Identifying mRNA expression patterns and correlating with relevant clinical parameters, identifying translocation or rearrangement events.
- Copy number and tumor purity - Clustering cases according to copy number alteration or loss-of-heterozygosity events, identifying candidate drivers of copy number alterations, estimating tumor purity of the samples.
- miRNA analysis - Analyzing miRNA expression to correlate with patterns of mRNA expression and identify expression regulation networks, correlating miRNA data with relevant clinical parameters.
- Long non coding RNA (lnRNA) – Analyzing lnRNA expression patterns and correlating with patterns of mRNA expression or expression regulation networks.
- Batch effects and data integration – Identifying batch effects that might have been accrued during processing of samples, devising bioinformatics methods to correct such effects, determining biologically relevant groups that can subsequently be analyzed in the context of clinical data.
- Methylation analysis – Identifying DNA methylation patterns of interest and correlating patterns with relevant clinical parameters, correlating patterns with mRNA expression data to propose gene regulation mechanisms.
- Pathway analysis – Identifying biological pathways that have been altered, performing multi-omic data analyses to identify altered pathways and potential clinical relevance.
- Single cell RNA sequencing – Identifying cell clusters according to gene expression patterns, extracting expression levels and correlating with relevant clinical parameters, identifying translocation/rearrangement events, and identifying cell clusters or subclones of interest.
- Circulating cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA) – Analyzing “liquid biopsies,” or blood samples for cfDNA or ctDNA to establish correlations between mutations in tumor tissue and ctDNA, developing methods to utilize the technology as a diagnostic and prognostic tool, and creating models of disease burden and progression in cancer development.
- Long-read sequencing – Assembling genomes, identifying structural variants, sequencing through repetitive regions, phasing critical variants.
- Spatial genomics – Analyzing gene expression data with spatial information, produced from different emerging spatial genomics platforms.
- Digital Imaging – Mining histopathology images for elements that aid in diagnostic or prognostic efforts, applying machine learning to learn relevant features.
New Molecular Profiling Platforms to Explore New Facets of Cancer
OCG continues to expand and develop new genomic data and analysis resources for the cancer research community. Through the GDAN and other OCG programs, OCG is exploring new ways to mine the data and learn new things about cancer from the massive dataset.
Additionally, as new molecular platforms become available, OCG explores utilizing these platforms to complement existing datasets. New platforms may be utilized in new structural genomics projects or in some cases to further characterize existing samples. Existing or new Genome Characterization Centers may be sought out to provide these capabilities. These newer platforms include:
- Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
- Single-cell RNA
- Single-cell DNA
- Spatial genomics
OCG considers how these new technologies may be applied to enhance what we can learn about cancer. For example, can single-cell or spatial technologies provide much needed insights into the tumor microenvironments of tumors that don’t respond to treatment? How can the technologies be used to further what we can learn from TCGA or other existing datasets?
For example, GDAN researchers applied the ATAC-seq chromatin accessibility assay to 410 TCGA tumor samples, getting an unprecedented systematic look at gene dysregulation in cancer. With this low-cost assay, the researchers were able to discover new DNA regulatory elements and a new class of mutations falling within these elements that may play a key role in cancer.
In addition to applying new molecular platforms to TCGA samples, OCG is also working to perform whole-genome sequencing for the complete set of TCGA samples. These rich datasets, along with analyses and methods developed by the GDAN, could help facilitate the discovery of new diagnostic and prognostic markers, new targets for pharmaceutical interventions, and new cancer prevention and treatment strategies.
Current GDAN Centers
The GDAN is comprised of individual Genome Data Analysis Centers (GDACs), each contributing distinct functions, capabilities, and analytical components. Each GDAC works collaboratively within the network and also with other components of OCG’s Genome Characterization Pipeline. The current GDACs and their area of expertise in computational genomics are described below.
| Center, PIs | Description |
|---|---|
Weill Medical College of Cornell University Olivier Elemento, Nicolas Robine, Cora Sternberg | The joint WCM-NYGC Center for Functional and Clinical Interpretation of Tumor ProfilesThe joint Weill Cornell Medicine-New York Genome Center (WCM-NYGC) Center for Functional and Clinical Interpretation of Tumor Profiles will perform integrative analyses of coding and non-coding variants to detect and unravel the function of specific classes of mutations and assess their clinical potential. Specialties: DNA mutations, copy number and purity analysis |
| Washington University Li Ding, Ramaswamy Govindan | Deep exploration of drivers, evolution, and microenvironment toward discovering principal themes in cancerUnderstanding the whole spectrum of inherited and acquired genetic changes using established and emerging technologies will lead to effective diagnosis and treatment strategies for each patient's cancer. We undertake this work using a comprehensive suite of established bioinformatics tools to examine mutations and germline predisposition, tumor cell populations, evolution, and the tumor microenvironment; and integrate these dynamics into larger themes in cancer. Specialties: DNA mutations, long-read sequence analysis, scRNA-Seq analysis, and spatial genomics data analysis (with connection to digital imaging analysis) |
| University of Texas MD Anderson Cancer Center Rehan Akbani, Bradley Broom, John Weinstein | A Genome Data Analysis Center Focused on Batch Effect Analysis and Data IntegrationThe principal goals of the Genome Data Analysis Center (GDAC) proposed here are (i) to perform batch effects identification, quantitation, diagnosis, and (when appropriate) correction, proactively and as requested by the Network; (ii) to contribute when useful to integrated cluster analysis, RPPA proteomic data analysis, and high- level interactive visualization of omic data (including single-cell sequencing data); (iii) to communicate results to other network members, project stakeholders, and the scientific community. Specialties: Batch effects, integration |
| Princeton University Benjamin Raphael, Jen Jen Yeh | Pathway, Network and Spatiotemporal Integration of Cancer Genomics DataOur center will develop and apply novel computational approaches to identify biological pathways that are altered in cancer and to characterize tumor heterogeneity, suggesting new approaches for cancer diagnosis and treatment. Specialties: Pathway analysis, integration |
| Oregon Health & Science University Paul Spellman | OHSU Center for Specialized Data Analysis as part of the GDANThe GDAN represents an attempt to bring retrospective precision medicine to the NCI's clinical trial infrastructure. As such it is a great opportunity to learn why trials that have occurred worked at a broad level, or identify patients who likely benefited from therapy, even when the trials were not successful. Our participation in this network will bring the most robust approaches for mutation calling and expression analysis, will bring novel pathway analysis approaches, and will bring an analysis of tumor genetic heterogeneity and evolution. Specialties: DNA mutation, expression, pathway analysis, single cell RNA-seq |
University of California Santa Cruz Josh Stuart | UCSC-Buck Genome Data Analysis Center for the Genomic Data Analysis Network v2.0Our proposal would contribute to the future GDAN to apply computational approaches that unite these data with the growing information contained in new single-cell datasets to provide key information about the types of cells that comprise a tumor and its microenvironment through the use of mRNA gene expression signatures. We will share new results of the GDAN through the TumorMap and Xena Browsers to support exploration of the data through collaborations with Analysis Working groups and work with the consortium to define state-of-the-art machine-learning methods to predict therapy response in new clinical trial datasets. Specialties: Pathway analysis, integration, expression, DNA mutations |
| Broad Institute Inc. Rameen Beroukhim | Center for the Comprehensive Analysis of Cancer Somatic Copy-Number Alterations, Rearrangements, and Long-Read Sequencing DataSomatic copy-number alterations (SCNAs) and the rearrangements that generate them are, together with mutations, the major somatic genome alterations in human cancer, and understanding them yields insights into how to diagnose and treat cancers. We have extensive experience in developing methods to detect and interpret SCNAs and rearrangements and have applied these methods across tens of thousands of tumors, discovering new molecular subtypes of cancer that may benefit from new therapeutic strategies. We propose to renew our Genomics Data Analysis Center that will specialize in conducting state-of-the-art analyses of SCNAs and rearrangements from both short read and long read sequencing data to answer clinically and biologically relevant questions that are tailored to the needs of the wider Genomics Data Analysis Network. Specialties: Copy number, single cell RNA-seq, circulating cell free DNA |
| Broad Institute, Inc. Gad Getz, Esther Rheinbay | Comprehensive analysis of point mutations in cancerWith expertise in discovering and characterizing point mutations in the cancer genome, we aim to integrate our state-of-the-art rigorous tools and pipelines for robust point mutation characterization and driver discovery into the Genome Data Analysis Network (GDAN) alongside other GDACs with expertise in complementary fields. Understanding the significance of the point- mutation drivers in the context of analyses from other GDACs in the GDAN will provide the cancer field with a deep, comprehensive understanding of many cancer types that can inform future clinical therapeutics and advance precision medicine at the single-patient level. Specialties: DNA mutations, cell free circulating DNA, expression, single cell RNA-seq, pathway analysis |
| Van Andel Research Institute Peter Laird, Hui Shen | Integrative Cancer Epigenomic Data Analysis Center (ICE-DAC)Cancer arises from the loss of control of growth in cells. Although attention has focused mostly on genetic mutations as the origin of these defects, it has become clear that changes in the epigenetic control of gene activity also contribute to this loss of growth control. This project aims to provide epigenetic analysis of clinical trial epigenomic data to advance our understanding of how epigenetic defects impact cancer biology and clinical outcome. Specialties: Methylation |
| University of North Carolina Chapel Hill Katherine Hoadley, David Hayes | Specialized RNA analysis center for integrative genomic analysesLarge scale, multi-platform genomic projects in translational studies are often difficult to accomplish by individual laboratories. Collaborate team science networks, like The Cancer Genome Atlas and the Genome Data Analysis Network, bring together researchers with varied expertise to generate, analyze, and provide the data back to the research community for larger use and impact. We propose to provide expertise in RNA sequencing, spatial genomics, data integration, and clinical outcomes analysis as a Genome Data Analysis Center in support of the Genome Data Analysis Network. Specialties: Expression, spatial genomics, integration |
| Sloan-Kettering Institute for Cancer Research Nikolaus Schultz, Sohrab Shah | The MSK Genomic Data Analysis Center for Tumor EvolutionThe MSK Genomic Data Analysis Center for Tumor Evolution will create a software platform for analysis of DNA mutations in cancer to help researchers and clinicians better understand why cancers often relapse. As cancer is a disease that changes at the cellular level over time, with some cells killed by treatment while others survive, we need to understand which mutations lead to treatment failure in specific patients. We expect that with improved tools that can measure, monitor and interpret changes in disease over time, we will make advances that allow for better management of cancer and prevention of relapse. Specialties: DNA mutation, spatial genomics, single cell RNA-seq |