RAS Initiative Biochemistry & Biophysics Research
The RAS Initiative Biochemistry and Biophysics Research Team uses a variety of biochemical and biophysical assays to characterize the interaction between RAS proteins and their effectors in solution or on the membrane. In addition, our researchers develop assays to identity small molecules inhibitors of RAS.
Progress
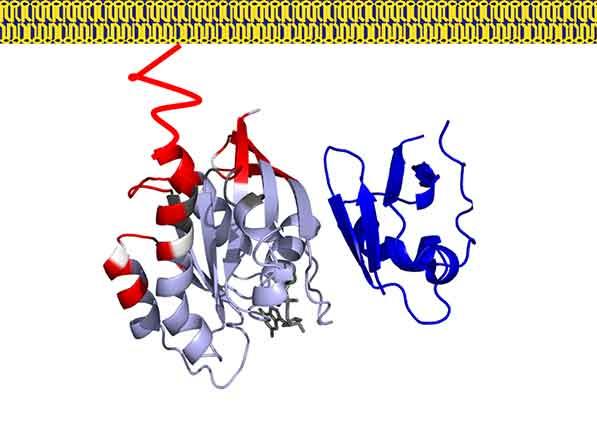
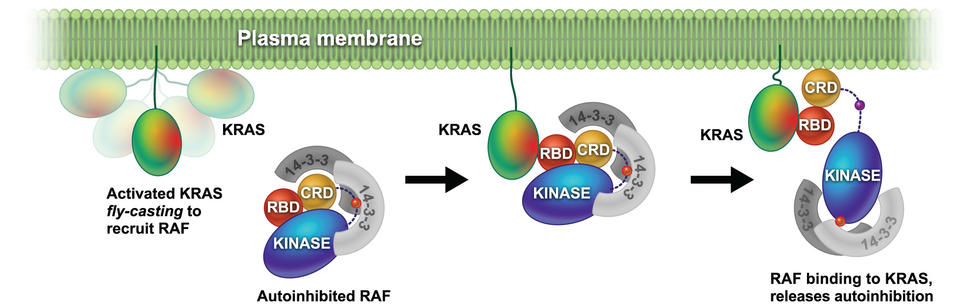
In our previous work we have used a combination of NMR, neutron reflectivity, protein foot printing and SPR to determine the interaction of KRAS with the domains of RAF and other effector proteins on membrane mimetics.
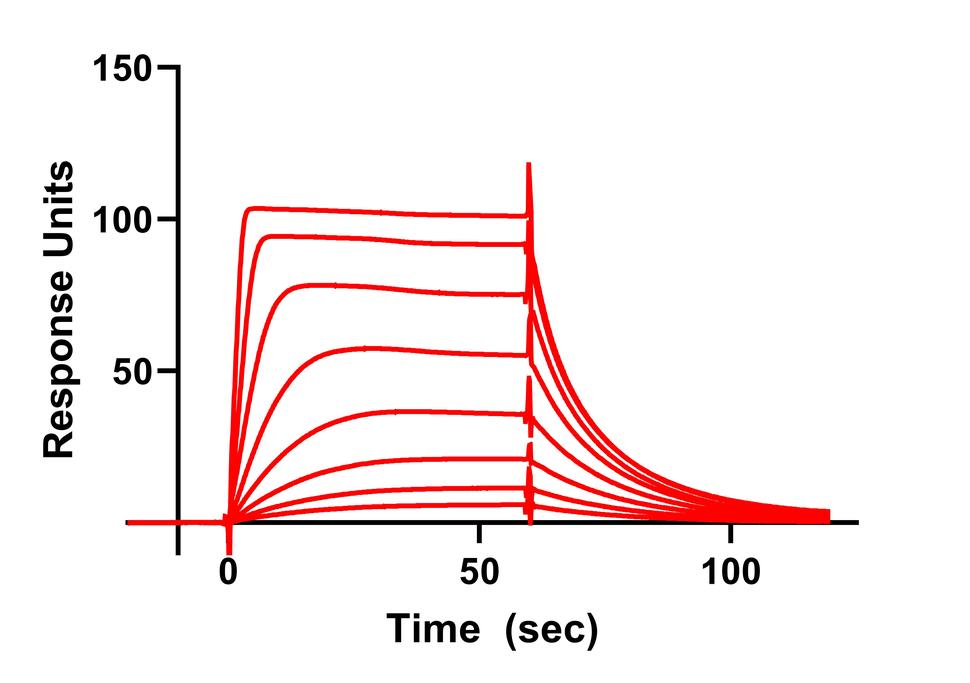
Currently we have assays that measure the interaction between RAS and effectors (RAF, PI3K, RAL GDS), GTPase Activating Proteins and Guanine Nucleotide Exchange Factors. In addition, we use SPR and MST to validate the binding of small molecule inhibitors of RAS.
Projects
We are constantly evaluating new approaches to target KRAS and working with our colleagues to screen new molecules for their inhibitory activity towards KRAS.
In addition, we are focused on understanding how RAS binding and membrane engagement mediate the activation of RAF. We are using a combination of biophysical methods and computational simulations to shed light on the process of RAS mediated RAF activation.
Tools
With our specialized tools, we measure changes in the biophysical properties (fluorescence, temperature, refractive index, and electro-magnetic) of recombinant proteins to quantitate with interaction with small molecules or other proteins.
- Surface Plasmon Resonance Spectroscopy
- Proximity assays: Alpha and HTRF
- Fluorescence-based activity assays
- Thermal shift assays
- Microscale Thermophoresis
- Solution state NMR
- Neutron reflectivity
The RAS Initiative Biochemistry and Biophysics Research Team
Team lead and contact
Andy Stephen
stephena@nih.gov
301-846-1634
Collaborators
We collaborate with investigators who provide expertise in biophysical methodologies such as NMR (Marco Tonelli), EPR (Jason Sidabras), neutron reflectometry (Frank Heinrich), neutron scattering (Oak Ridge National Laboratory) and protein:membrane interactions (Stephen Sligar). In addition, we complement our experimental measurements with molecular dynamics simulations of KRAS and effector interaction on membranes (Lawrence Livermore National Laboratory and Los Alamos National Laboratory).
- Theras
- Sanofi
- Frank Heinrich, NIST Center for Neutron Science
- Marco Tonelli, National Magnetic Resonance Facility at Maddison
- Jason Sidabras, Medical College of Wisconsin
- Stephen Sligar, University of Illinois Champagne Urbana
- Lawrence Livermore National Laboratory
- Los Alamos National Laboratory
- Oak Ridge National Laboratory
Publications
- Heinrich F, Van QN, Jean-Francois F, Stephen AG, Lösche M. Membrane-bound KRAS approximates an entropic ensemble of Configurations. Biophysical Journal. 2021;120(18):4055-4066. doi:10.1016/j.bpj.2021.08.008 [PubMed Abstract]
- López CA, Agarwal A, Van QN, Stephen AG, Gnanakaran S. Unveiling the dynamics of kras4b on lipid model membranes. The Journal of Membrane Biology. 2021;254(2):201-216. doi:10.1007/s00232-021-00176-z [PubMed Abstract]
- Van QN, Prakash P, Shrestha R, Balius TE, Turbyville TJ, Stephen AG. Ras nanoclusters: Dynamic signaling platforms amenable to therapeutic intervention. Biomolecules. 2021;11(3):377. doi:10.3390/biom11030377 [PubMed Abstract]
- Van QN, López CA, Tonelli M, et al. Uncovering a membrane-distal conformation of KRAS available to recruit RAF to the plasma membrane. Proceedings of the National Academy of Sciences. 2020;117(39):24258-24268. doi:10.1073/pnas.2006504117 [PubMed Abstract]
- Travers T, López CA, Agamasu C, et al. Anionic lipids impact RAS-binding site accessibility and membrane binding affinity of CRAF RBD-CRD. Biophysical Journal. 2020;119(3):525-538. doi:10.1016/j.bpj.2020.06.021 [PubMed Abstract]
- Tran TH, Alexander P, Dharmaiah S, et al. The small molecule BI-2852 induces a nonfunctional dimer of Kras. Proceedings of the National Academy of Sciences. 2020;117(7):3363-3364. doi:10.1073/pnas.1918164117 [PubMed Abstract]
- Agamasu C, Ghirlando R, Taylor T, et al. Kras prenylation is required for bivalent binding with calmodulin in a nucleotide-independent manner. Biophysical Journal. 2019;116(6):1049-1063. doi:10.1016/j.bpj.2019.02.004 [PubMed Abstract]
- Esposito D, Stephen AG, Turbyville TJ, Holderfield M. New weapons to penetrate the armor: Novel reagents and assays developed at the NCI RAS initiative to enable discovery of Ras Therapeutics. Seminars in Cancer Biology. 2019;54:174-182. doi:10.1016/j.semcancer.2018.02.006 [PubMed Abstract]