CytoScreen: A Miniaturized Imaging Platform to Develop Combinatorial Therapies
, by Tania Q. Vu, Ph.D. and Thomas Jacob, Ph.D.
Hematologic malignancies represent the fifth most common type of cancer and fourth most common form of cancer-related death. More than 1 million people in the U.S. suffer from a hematologic malignancy; every year, another 150,000 patients are diagnosed with lymphoma, leukemia or myeloma. Targeted therapies have been a recent focus of drug development, but the majority of patients eventually develop resistance even to these new drugs. As a result, many leukemia patients eventually succumb to their diseases due to resistance to both conventional chemotherapies and newer targeted agents. Hence, there is an urgent need to better understand the pathways underlying drug resistance and to identify novel drugs or combinations of drugs that can effectively inhibit these pathways.
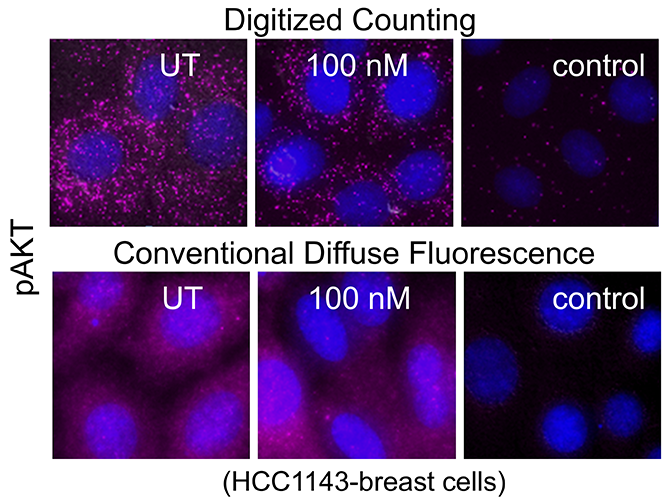
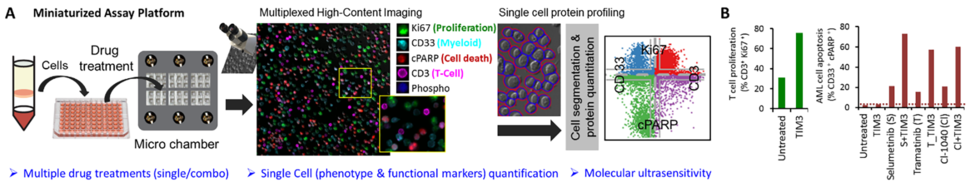
Major efforts are underway to develop combinatorial therapies that can target multiple biological pathways for effective synergistic cell killing that can help decrease the risk of cancer relapse. A major technical challenge is the lack of screening platforms that allow assessment of optimal combinations of drug conditions directly in primary patient cells. Flow cytometry is a widely-used option, however the amount of sample required limit the number of drug combinations that can be tested. Other ex vivo screening platforms lack single cell granularity and hinder the detection of cells that are resistant to drug treatments. Moreover, many protein targets (e.g. phosphoproteins, immune modulatory molecules induced via inflammation) may often be present at low levels. Hence, negative detection cannot be conclusively interpreted, and it is difficult to discriminate the signal from diffuse background noise. To address these issues, Drs. Tania Vu and Thomas Jacob at the Knight Cancer Center, Oregon Health and Science University (OHSU) have developed a single cell imaging platform, the CytoScreen. The CytoScreen is a miniaturized assay platform performed in multi-well imaging chambers that allows testing of drug combinations in primary patient cells with single cell granularity and at high detection sensitivity. In this assay, protein levels are quantified by digitized, discrete counting of fluorophore-tagged proteins. This molecular digital detection imparts amplification-free detection sensitivity and provides significant improvement in protein quantification over current methods that quantify the total diffuse fluorescence per cell1. This is an ultrasensitive system and scales down the amount of sample required to perform the assay in patient samples. This assay offers new capabilities for profiling the therapeutic effect of multiple protein targets in different cell subpopulations that were previously challenging to measure.
In collaboration with Drs. Jeffrey Tyner, Evan Lind, Brian Druker and others at the OHSU’s Knight Cancer Center and Dr. Jody Martin at Becton Dickinson, the team has customized CytoScreen assays for the identification of effective new treatment options – especially those that address combination immunotherapies and drug-resistance. This assay allows testing a panel of drug combinations with multi-parameter readouts of T-cell activation and tumor-cell killing as well as readouts of stem/progenitor cell phosphoactivation with single cell granularity. The system has been used to assess drug response from acute myeloid leukemia (AML), chronic myeloid leukemia (CML), and chronic lymphocytic leukemia (CLL) patients.
The Beat AML study is a groundbreaking collaborative clinical study characterizing the molecular characteristics and drug exposure response for clinically annotated adult AML cases2. The Beat AML is the world’s largest AML cohort, and the goal of this study is to evaluate investigational or combinatorial therapies to advance precision oncology. The CytoScreen assay system is adapted to identify small molecule/immune checkpoint combinations that can operate synergistically to boost anti-tumor immune response and kill tumor cells in AML samples. The combinations of small molecule/immune checkpoint inhibitors that can overcome resistance are computationally predicted using RNA-seq and ex vivo small molecule sensitivity data from the Beat AML cohort. This initial screen showed that mucin-domain containing protein 3 (TIM3) and mitogen-activated protein kinase kinase (MEK) inhibitors can have synergistic effects on T-cell immunoglobulins. The CytoScreen studies also showed that in certain patients, TIM3 in combination with certain MEK inhibitors can elicit more effective T-cell proliferation than the single immune checkpoint agent alone.
New technical capabilities – including increased multiplexing, profiling function and pathway activity not only of single leukemic cells but also T-cells and stem cell subpopulations as well as directly in individual patient samples – are currently in progress. Cancer Target Discovery and Development (CTD²) Center at OHSU is using this platform to screen combinations of targeted pathways and immune-modulating therapies in hematologic malignancies. The team at the Knight Cancer Research Center is optimistic that the integrations of technologies like this, along with other ex vivo functional technologies and predictive computation tools, will enable them to uncover effective drug combinations that can be advanced to clinical trial testing to improve therapy not only for AML and CLL patients but also for other leukemias in the near future.
In conclusion, the CytoScreen is a new assay platform that addresses issues in sample requirement and molecular detection sensitivity. This assay facilitates routine assessment of single-cell drug target effects in most leukemia patient samples and provides multiplexed readouts of specific cell subpopulations (T- cells, stem cells, leukemic cells) as well as their functional status (death, phosphoactivity, proliferation state) with quantitative accuracy.
References
- Jacob T, Agarwal A, Ramunno-Johnson D, et al. Ultrasensitive proteomic quantification of cellular signaling by digitized nanoparticle-protein counting. Scientific Reports. 2016 June 20;(6):28163. (PMID: 27320899)
- Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukemia. Nature. 2018 October;562(7728):526-531. (PMID: 30333627)